Our lab studies eukaryotic gene expression. We are concentrating on three areas: (A) the functions and interactions of the RNA polymerase II (RNApII) basal transcription factors, (B) the communication between chromatin and the transcription machinery, and (C) mRNA processing enzymes and their interactions with RNApII. Using the yeast Saccharomyces cerevisiae, a combination of biochemical and genetic techniques are being brought to bear on these questions. Several dozen proteins are required simply to initiate transcription, and many more are required for processes linked to transcription. Therefore, it is now necessary to decipher the functions of each of the individual factors. Some of our recent projects:
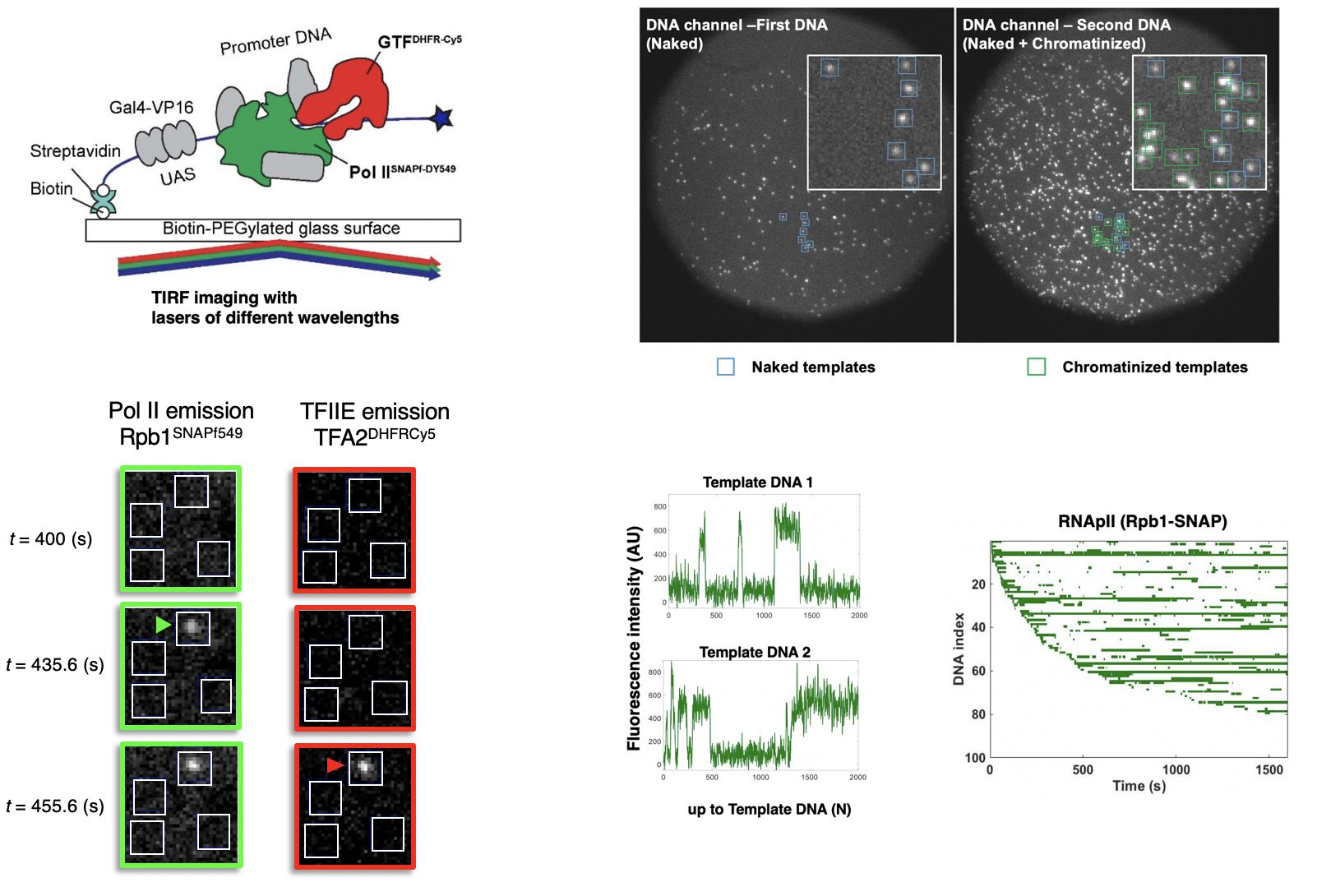
This project traces back to Steve's PhD work on TATA-Binding Protein (Buratowski et al., Nature 1988) and analysis of RNA polymerase II initiation complexes (Buratowski et al., Cell 1989). Our recent experiments use mass spectrometry and single molecule microscopy to understand the dynamics of transcription activation and initiation. Current topics include:
- Dynamics of transcription coactivators
- Understanding the relationship between TBP and TAFs in TFIID
- Assembly pathway of initiation complexes
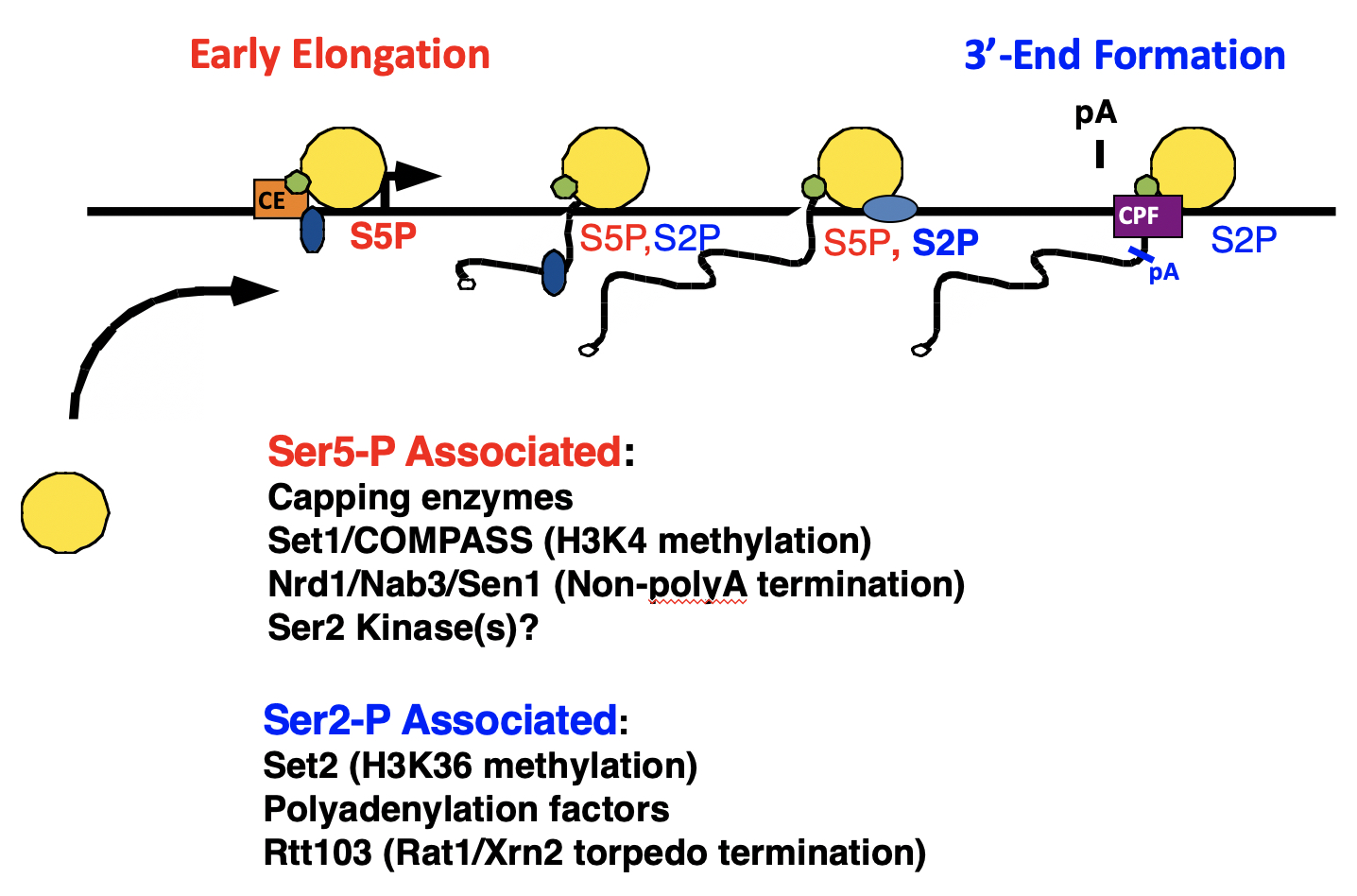
The RNApII C-terminal domain (CTD) and mRNA processing enzymes. mRNAs are capped at the 5' end and polyadenylated at the 3' ends. Our lab discovered the CTD Code, where the pattern of CTD phosphorylation changes at various points of transcription initiation and elongation. These different phosphorylated forms bind different sets of factors involved in regulation of elongation, termination, capping, splicing, and polyadenylation.
Current projects in the lab include:
- Development of an in vitro system that reproduces the CTD cycle
- Direct mass spectrometry of CTD phosphorylation
- The role of CTD kinases and phosphatases
- Interaction of capping enzyme with CTD
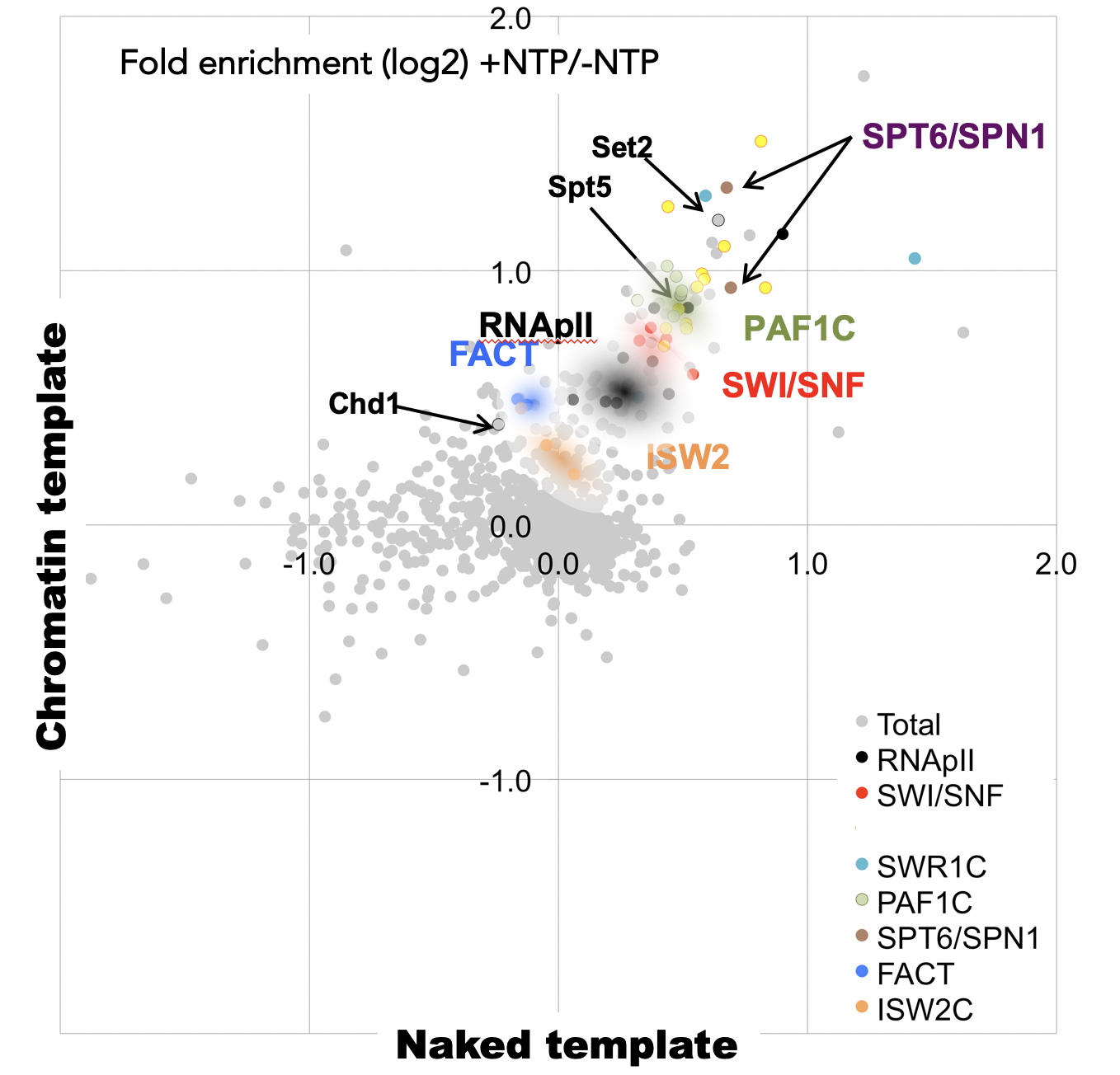
We are studying the many factors that modulate transcription elongation and termination by RNApII. We have found that different mechanisms are used for termination at different classes of genes. Genes that encode polyadenylated mRNAs use an exonuclease-dependent pathway often called the "torpedo model" (Kim et al. Nature 2004), while genes for the non-polyadenylated sn/snoRNAs use a pathway that includes the exosome and the Nrd1 and Nab3 RNA binding proteins (Kim et al, Mol. Cell 2006). We are working to further understand the two pathways and how the choice is made between them. We have also developed a system for analyzing elongation complexes in vitro using quantitative mass spectrometry (Joo et al., Genes Dev. 2019).
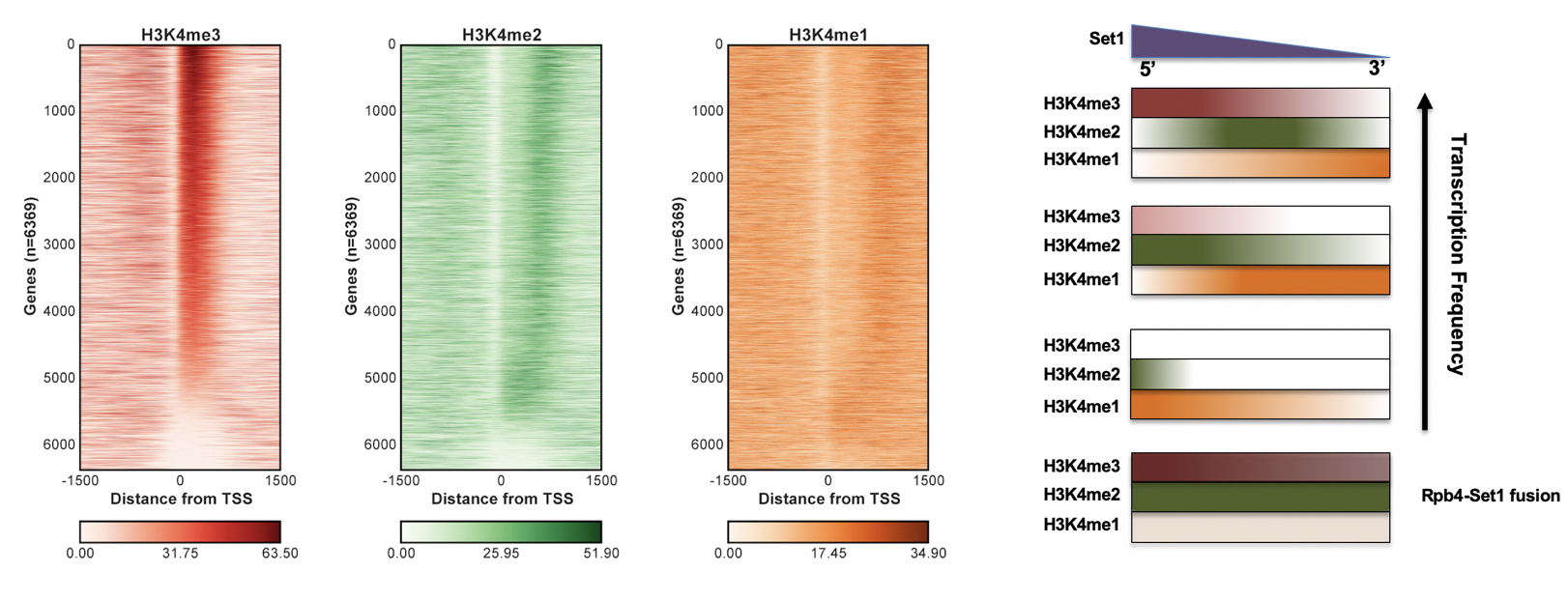
We and others showed that the act of transcription causes major changes in the nucleosomes that package the gene. In particular, the histone methyltransferases Set1 and Set2 are targeted to promoter and coding regions, respectively, via binding to the phosphorylated RNApII CTD. We discovered that these transcription-coupled histone methylation patterns target specific histone deacetylases (Keogh et al., Cell 2005; Kim and Buratowski, Cell 2009). Surprisingly, many mRNA promoters are overlapped by non-coding RNA transcription, and the interplay between histone modifications generated by the coding and non-coding transcription is used to modulate gene expression (Kim et al., Cell 2012). Many of these co-transcriptional histone modifiers have been linked to human cancers, but yeast provides a perfect model system for getting at their basic functions.